Estimate organic carbon from organic matter values
Usage
estimate_oc(
df = NULL,
core = "core",
site = "site",
ecosystem = "ecosystem",
species = "species",
om = "om",
oc = "oc"
)Arguments
- df
A tibble or data.frame containing all the data. Must have at least five columns (see arguments below).
- core
Character Name of the column with the id of the core to which the sample belongs
- site
Character Name of the column reporting sample site.
- ecosystem
Character Name of the column reporting ecosystem type. To apply published equations for OC estimation, ecosystem names should be either "Salt Marsh", "Seagrass" or "Mangrove".
- species
Character Name of the column reporting the main species in the site.
- om
Character Name of the column reporting organic matter values.
- oc
Character Name of the column reporting organic carbon values.
Value
The initial tibble or data.frame with three new columns:
one column with estimated organic carbon values (eOC) in %
the standard error of the prediction (eOC_se)
the type of model used for estimation (origin)
In addition, a plot with the relationship between organic matter and estimated organic carbon values.
Details
Estimation of organic Carbon is done by means of linear regressions on
log(organic carbon) ~ log(organic matter), which return estimated organic carbon
value for each organic matter value provided. If there is a value for organic carbon
for that sample it returns the same value; otherwise, it estimates organic carbon
from a model fitted to that site, or a model fitted to that species, or else
a model fitted to that ecosystem. If there are too few samples (<10) to build a
reliable model or the model fit is too poor (r2 < 0.5), estimate_oc() uses the equations
in Fourqurean et al. (2012) doi:10.1038/ngeo1477
for seagrasses,
Maxwell et al. (2023) doi:10.1038/s41597-023-02633-x
for salt marshes
and Piñeiro-Juncal (in prep.) for mangroves to estimate the organic carbon.
Examples
bluecarbon_decompact <- decompact(bluecarbon_data)
#> Warning: Setting compaction = 0 for these cores: Sm_03_04, Sg_10_02, Sg_11_03, Sm_05_01, Sm_06_01
out <- estimate_oc(bluecarbon_decompact)
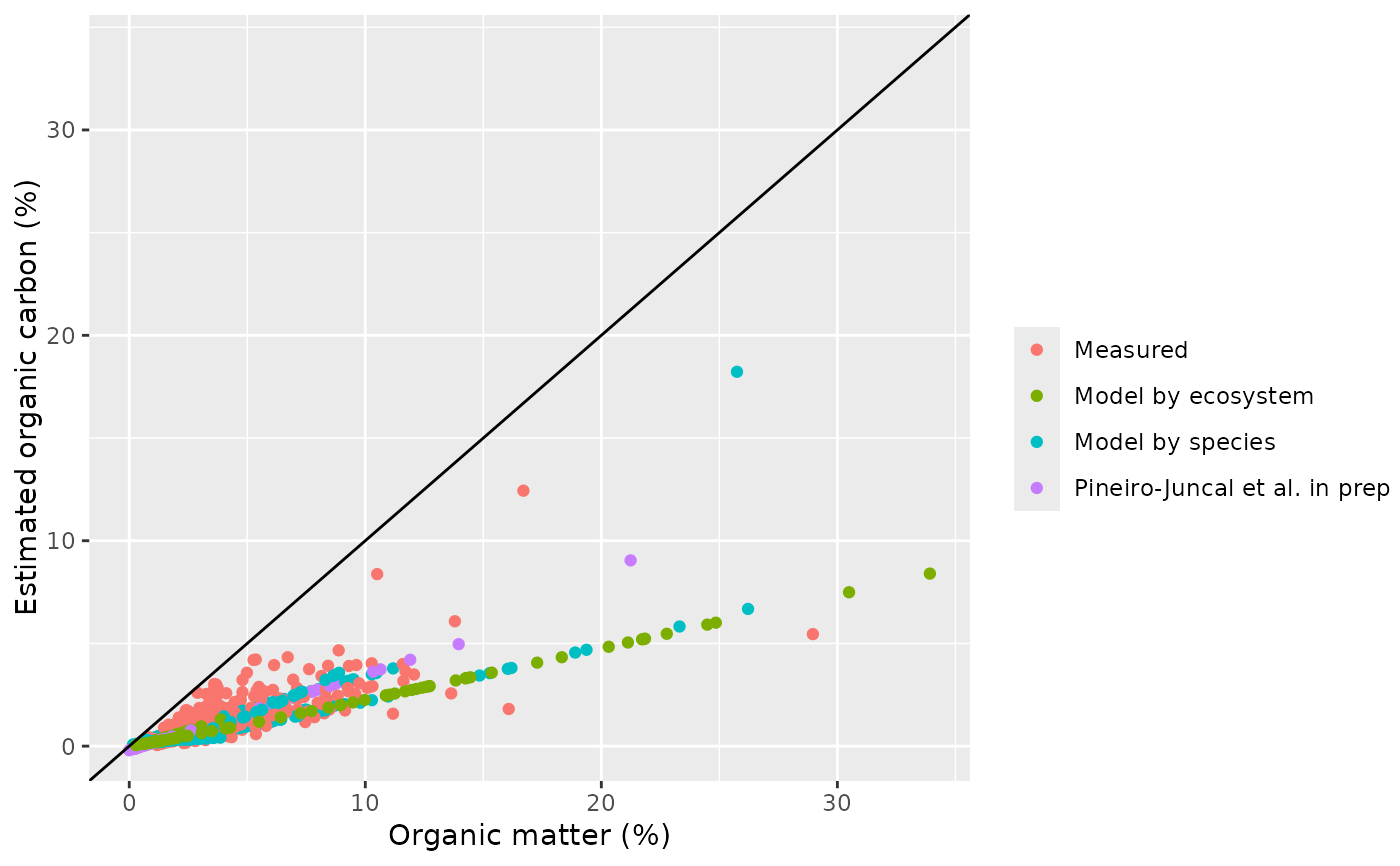 #> Warning: The following cores had samples with organic carbon values below the organic carbon range used to built the model: Sg_04_01, Sm_04_03, Sm_04_04, Sm_05_01
#> Warning: The following cores had samples with organic carbon values above the organic carbon range used to built the model: Sg_04_01, Sm_03_01, Sm_04_02, Sm_04_03, Sm_04_04, Sm_05_01
head(out$data)
#> site core ecosystem species compaction mind maxd dbd
#> 1 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 0 1 0.7352912
#> 2 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 1 2 0.9754336
#> 3 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 2 3 0.8698411
#> 4 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 3 4 1.0272564
#> 5 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 4 5 0.9307887
#> 6 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 5 6 1.4696196
#> om oc age mind_corrected maxd_corrected eoc eoc_se
#> 1 6.554329 NA 8 0.000000 1.060606 2.296034 0.04585142
#> 2 NA NA 13 1.060606 2.121212 NA NA
#> 3 7.382634 NA 15 2.121212 3.181818 2.566545 0.05166124
#> 4 NA NA 22 3.181818 4.242424 NA NA
#> 5 8.026646 NA 29 4.242424 5.303030 2.775515 0.05591166
#> 6 NA NA 35 5.303030 6.363636 NA NA
#> origin
#> 1 Model by species
#> 2 <NA>
#> 3 Model by species
#> 4 <NA>
#> 5 Model by species
#> 6 <NA>
out$models
#> $Mangrove
#> $Mangrove$ecosystem_model
#> NULL
#>
#> $Mangrove$multispecies_model
#> NULL
#>
#> $Mangrove$site_models
#> NULL
#>
#>
#> $`Salt Marsh`
#> $`Salt Marsh`$ecosystem_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.667 1.077
#>
#>
#> $`Salt Marsh`$multispecies_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r) * species_r, data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.9154 1.1678
#> species_rSpartina maritima log(om_r):species_rSpartina maritima
#> 1.0662 -0.4614
#>
#>
#> $`Salt Marsh`$site_models
#> $`Salt Marsh`$site_models$Sm_01
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.8492 0.7064
#>
#>
#> $`Salt Marsh`$site_models$Sm_02
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -2.281 1.484
#>
#>
#> $`Salt Marsh`$site_models$Sm_03
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.892 1.121
#>
#>
#>
#>
#> $Seagrass
#> $Seagrass$ecosystem_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.463 1.278
#>
#>
#> $Seagrass$multispecies_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r) * species_r, data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.5655 0.5137
#> species_rPosidonia oceanica species_rZostera noltii
#> 0.6371 -0.5090
#> log(om_r):species_rPosidonia oceanica log(om_r):species_rZostera noltii
#> 0.4222 1.0185
#>
#>
#> $Seagrass$site_models
#> $Seagrass$site_models$Sg_01
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.9583 1.3323
#>
#>
#> $Seagrass$site_models$Sg_02
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.9599 1.2759
#>
#>
#> $Seagrass$site_models$Sg_03
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -2.861 2.589
#>
#>
#> $Seagrass$site_models$Sg_04
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.808 1.346
#>
#>
#> $Seagrass$site_models$Sg_05
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.5655 0.5137
#>
#>
#> $Seagrass$site_models$Sg_06
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.8815 0.8568
#>
#>
#>
#>
#> Warning: The following cores had samples with organic carbon values below the organic carbon range used to built the model: Sg_04_01, Sm_04_03, Sm_04_04, Sm_05_01
#> Warning: The following cores had samples with organic carbon values above the organic carbon range used to built the model: Sg_04_01, Sm_03_01, Sm_04_02, Sm_04_03, Sm_04_04, Sm_05_01
head(out$data)
#> site core ecosystem species compaction mind maxd dbd
#> 1 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 0 1 0.7352912
#> 2 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 1 2 0.9754336
#> 3 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 2 3 0.8698411
#> 4 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 3 4 1.0272564
#> 5 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 4 5 0.9307887
#> 6 Sg_01 Sg_01_01 Seagrass Posidonia oceanica 5.714286 5 6 1.4696196
#> om oc age mind_corrected maxd_corrected eoc eoc_se
#> 1 6.554329 NA 8 0.000000 1.060606 2.296034 0.04585142
#> 2 NA NA 13 1.060606 2.121212 NA NA
#> 3 7.382634 NA 15 2.121212 3.181818 2.566545 0.05166124
#> 4 NA NA 22 3.181818 4.242424 NA NA
#> 5 8.026646 NA 29 4.242424 5.303030 2.775515 0.05591166
#> 6 NA NA 35 5.303030 6.363636 NA NA
#> origin
#> 1 Model by species
#> 2 <NA>
#> 3 Model by species
#> 4 <NA>
#> 5 Model by species
#> 6 <NA>
out$models
#> $Mangrove
#> $Mangrove$ecosystem_model
#> NULL
#>
#> $Mangrove$multispecies_model
#> NULL
#>
#> $Mangrove$site_models
#> NULL
#>
#>
#> $`Salt Marsh`
#> $`Salt Marsh`$ecosystem_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.667 1.077
#>
#>
#> $`Salt Marsh`$multispecies_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r) * species_r, data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.9154 1.1678
#> species_rSpartina maritima log(om_r):species_rSpartina maritima
#> 1.0662 -0.4614
#>
#>
#> $`Salt Marsh`$site_models
#> $`Salt Marsh`$site_models$Sm_01
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.8492 0.7064
#>
#>
#> $`Salt Marsh`$site_models$Sm_02
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -2.281 1.484
#>
#>
#> $`Salt Marsh`$site_models$Sm_03
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.892 1.121
#>
#>
#>
#>
#> $Seagrass
#> $Seagrass$ecosystem_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.463 1.278
#>
#>
#> $Seagrass$multispecies_model
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r) * species_r, data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.5655 0.5137
#> species_rPosidonia oceanica species_rZostera noltii
#> 0.6371 -0.5090
#> log(om_r):species_rPosidonia oceanica log(om_r):species_rZostera noltii
#> 0.4222 1.0185
#>
#>
#> $Seagrass$site_models
#> $Seagrass$site_models$Sg_01
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.9583 1.3323
#>
#>
#> $Seagrass$site_models$Sg_02
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.9599 1.2759
#>
#>
#> $Seagrass$site_models$Sg_03
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -2.861 2.589
#>
#>
#> $Seagrass$site_models$Sg_04
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.808 1.346
#>
#>
#> $Seagrass$site_models$Sg_05
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -1.5655 0.5137
#>
#>
#> $Seagrass$site_models$Sg_06
#>
#> Call:
#> stats::lm(formula = log(oc_r) ~ log(om_r), data = df)
#>
#> Coefficients:
#> (Intercept) log(om_r)
#> -0.8815 0.8568
#>
#>
#>
#>
